PyIntro for Researchers
Mohammad AlMarzouq
ISOM Department
Code Repository
Submit Your Research Papers
- Arab Journal of Administrative Sciences
- https://journals.ku.edu.kw/ajas
- Call for papers
- Call for reviewers
- Quick turnaround time
- Open access
- Counts towards your promotion at KU
Disclaimer
- This course will not make you an expert
- Gives you a lay of the land
- Proficiency comes with practice
- Assumes basic programming knowledge
Important Notes
- We issue commands in Terminal and Python Console
- Will identify commands with a
$or>>>prompt - You should not type the prompt
- If no prompt, then its python code
- Use in python prompt or editor
Using Terminal
- On VSCode, there is a terminal menu to open and use the terminal
- On Windows, you can use PowerShell or Command Prompt (cmd)
- On Mac, you can use Terminal
- On Linux, …. you shouldn’t be using linux if you don’t know how to use the terminal :)
- On Jupyter Notebook, use
!to run terminal commands within a cell
Outline
- Why Python?
- Why Not Python?
- Development Environment
- Hello World
- Basic Syntax
- Installing Libraries
- Important Libraries
- Introduction to Pandas
- Reproducible Research
- Final Thoughts
Why Python
- General purpose programming language
- Cross platform
- High adoption
- Especially in ML and Data Science
- Philosophy (Zen of Python)
- Documentation
- Thriving community
- Ecosystem
- Online resources and videos
Python Philosophy
>>> import this
Zen of Python
- Beautiful is better than ugly.
- Explicit is better than implicit.
- Simple is better than complex.
- Complex is better than complicated.
- Readability counts.
Zen of Python
- There should be one– and preferably only one –obvious way to do it.
- Now is better than never.
- Although never is often better than right now.
- If the implementation is hard to explain, it’s a bad idea.
- If the implementation is easy to explain, it may be a good idea.
- Namespaces are one honking great idea – let’s do more of those!
Why NOT Python
- Already invested in another language/tool
- Network
- Learning curve
- Not sure where to start
Development Environment
- Numerous tools and python distributions
- Can get overwhelming
- Required
- Python Interpreter to execute code
- Editor to write code
- Modern tools integrate both seamlessly
- Good news, you setup the environment once
Writing Code
- Any Text Editor, including notepad
- Alternatives provide more features
- Syntax Highlighting
- Code Completion
- Debugging
- Version Control
- Integration with other tools
Executing Code
- Local vs Cloud
- Execution Modes
- Script Mode
- Interactive Mode
- Hybrid/Notebook mode
Python Interpreters
- Some OSs come with Python pre-installed
- Python.org
- Most up to date
- Anaconda
- Bundled with many libraries for data science
- Easy to install and manage
Local Code Editors
- VSCode
- RStudio
- Jupyter Notebook
- Jupyter Lab
- Sublime Text
- Atom
- Notepad++
- Vim/Emacs
- PyCharm
- Spyder
Cloud Based Editors
- JupyterHub
- Github + mybinder
- Replit
- Kaggle
- Google Colab*
- CoCalc*
- Deepnote*
* Can be used to train and fine-tune DL models
IDE of Choice
- VSCode
- Likely to be part of your workflow for other languages/tasks
- Extensions and copilot make it a powerful tool
- Allows you to execute python in different modes, even notebooks
- Well integrated with Git/Github
- Replace
github.comwithgithub.devin the URL for any repo
- Replace
VSCode Requirements for Python
- Install Python Interpreter
- Install Python Extensions for VSCode
- Install Libraries as needed
Hello World
- Create a hello.py file
- Write the following code
print("Hello World")
- Run the code
Things to Know Before Starting
- Python Zen
- PEP8 and Idiomatic Python
- Writing and styling code the pythonic way
- Great summaries at
- Python is case sensitive
- Python 2 vs Python 3
Handy References
Overview of Basic Syntax
- Variables
- Data Types
- Operators
- Control Flow
- Functions
- Classes
- Collections
- Comprehensions
Variables
- A name that refers to a value
- No need to declare type
- Python is dynamically but strongly typed
- Recent versions have type hints
- Can be reassigned
Naming Conventions
- Lowercase
- Underscore to separate words
- Descriptive
- Avoid reserved words
Variable Assignment
x = 5
y = "Hello"
z = [1,2,3]
y = 10 # replaced value
Some Useful Functions
- print()
- input()
- type()
- dir()
- help()
- In notebooks, use
?or??after a function or variable- ?? will show the source code
Data Types
- int : 1, 2, 3
- float : 1.0, 2.0, 3.0
- str : “Hello”, ‘World’, “1”, “2”
- bool : True, False
- None
Collections
- list : [1,2,3]
- tuple : (1,2,3)
- set : {1,2,3}
- dict : {“a”:1, “b”:2, “c”:3}
Type Conversion
- int()
- float()
- str()
- bool()
- list()
- tuple()
- set()
- dict()
Type Conversion Example
x = "5"
y = int(x)
z = float(x)
- Does x equal y?
- Will throw an error if value cannot be converted
Arithmetic Operators
| Precedence | Operator | Description |
|---|---|---|
| 1 | () | Parentheses |
| 2 | ** | Exponentiation |
| 3 | +x, -x, ~x | Unary plus, Unary minus, Bitwise NOT |
| 4 | *, /, //, % | Multiplication, Division, Floor division, Modulus |
| 5 | +, - | Addition, Subtraction |
Bitwise Operators
| Precedence | Operator | Description |
|---|---|---|
| 6 | <<, >> | Bitwise shift operators |
| 7 | & | Bitwise AND |
| 8 | ^ | Bitwise XOR |
| 9 | | | Bitwise OR |
Comparison and Logical Operators
| Precedence | Operator | Description |
|---|---|---|
| 10 | ==, !=, <, <=, >, >= | Comparisons, Equality, Inequality |
| 11 | not | Logical NOT |
| 12 | and | Logical AND |
| 13 | or | Logical OR |
Creating Collections
x = []
y = [1,2,3]
z = [1,2,3 ,["foo", "bar"]]
Creating Collections
x = {}
y = set()
z = {"a":1, "b":2, "c":3}
k = {1,2,3,4,4,5}
Accessing Collections
- Indexing
- Slicing
- Iterating
Indexing
- Used to fetch a single element
- 0 based
- Negative indexing
- Out of range indexing
Example
x = [1,2,3,4,5]
print(x[0])
print(x[-1])
print(x[5])
Slicing
- Used to fetch a range of elements
- Start, Stop, Step
- Default values
- Negative slicing
Example
x = [1,2,3,4,5]
print(x[0:3])
print(x[:3])
print(x[3:])
print(x[::2])
print(x[::-1])
Iterating
- for loop
- while loop
- list comprehension
- generator expression
For Example
x = [1,2,3,4,5]
for i in x:
print(i) # this is a code block
Code Blocks in Python
- Some statements are followed by a code block
- Such statements end with a colon
: - Example: if, for, while, def, class, with, try, except, finally
- Code blocks are defined by indentation
- Indentation is typically 4 spaces
- Will get back to this later
Code Block Example
def function():
# do something
Another Example
if condition:
# do something
# do something else
if another_condition:
# do something
else:
# do something else
# do something else
Another Example
if condition:
# do something
# do something else
if another_condition:
# do something
else:
# do something else
else:
# do something else
# do something else
Another Example
for i in range(10):
# do something
# do something else
if i % 2 == 0:
# do something
else:
# do something else
if condition:
# do something
else:
# do something else
Back to Our For Example
x = [1,2,3,4,5]
for i in x:
print(i)
Another For Example
x = [1,2,3,4,5]
for i in range(len(x)):
print(x[i])
While Example
x = [1,2,3,4,5]
i = 0
while i < len(x):
print(x[i])
i += 1
Filtering Using For Loops
x = [1,2,3,4,5]
y = []
for i in x:
if i % 2 == 0:
y.append(i)
print(y)
List Comprehension
x = [1,2,3,4,5]
y = [i for i in x if i % 2 == 0]
print(y)
Comprehensions
- Exist for lists, sets, and dictionaries
- Used to create new collections from existing ones
- Can be used to filter, transform, or combine collections
- Can be nested
- More efficient than loops
Generator Expressions
- Similar to list comprehensions
- Use parentheses instead of square brackets
- Lazy evaluation
- More memory efficient
- Can be used to create infinite sequences
- Many functions in Python accept or return generator expressions
- Convert to list using
list()to evaluate - Otherwise, use in a for loop
Control Flow
- Conditional Statements
- if, elif, else
- loops
- for, while
If Statements
if condition:
# do something
elif condition: # optional
# do something else
else: # optional
# do something else
Conditions
- Comparison operators
==,!=,<,<=,>,>=
- Logical operators
and,or,not
- Membership operators
in,not in
- Identity operators
is,is not
- Truthy and Falsy values
Truthy and Falsy Values
| Value Type | Truthy | Falsy |
|---|---|---|
| Boolean | True | False |
| Null | None | |
| Zero | 0, 0.0, 0j | |
| Empty Collections | "", (), [], {}, set(), range(0) | |
| Others | Any other value not listed in Falsy |
Functions
- A block of code that only runs when it is called
- Can take arguments
- Mandatory or optional
- Positional or keyword
- Can return values
- Great for scoping variables
- Avoids global variables
- Makes code more modular
Void Function Example
def my_function():
print("Hello from a function")
- This is just a definition, the function is not executed
- To execute:
my_function()
Function with Arguments
def my_function(name):
print("Hello " + name)
Function with Return Value
def my_function(x):
return x**2
# execute
print(my_function(2))
Function with Default Argument
def my_function(x=2):
return x**2
# execute
print(my_function()) # Where is the argument?
Function with Keyword Arguments
def my_function(x=2, y=3):
return x**y
# execute
print(my_function(y=2, x=3))
# or
print(my_function(x=3, y=2))
Other Ways to Call The Functions
print(my_function(3, 2)) # positional
# or
print(my_function(x=3)) # keyword
# or
print(my_function(y=3))
# or
print(my_function())
What are the arguments?
Function with Variable Arguments
def my_function(*args):
return sum(args)
# execute
print(my_function(1,2,3,4,5))
print(my_function(1,2,3))
Function with Variable Keyword Arguments
def my_function(**kwargs):
return kwargs
# execute
print(my_function(a=1, b=2, c=3))
print(my_function(x=1, y=2))
Things to Note
argsandkwargsare just namesargsis a tuplekwargsis a dictionaryargsandkwargsare optional and must be at the endargs(positional) beforekwargs(keyword)
argsandkwargscan be used together
Variables Vs. Functions
- Variables store values
- Functions store instructions
- Both are considered variables that you can refer to
- Use parentheses to execute variables containing functions
What’s The Output?
print(x)
print("x")
print(input)
print(input())
Challenge
- Rename print to z
- Redefine print to be input
Solution
z = print
z("hello world")
print("Hello world")
print = input
print("Hello world")
Python Can Do Tricks
- But why is this useful?
- It means you can
- Pass functions as arguments to other functions
- You can return functions from other functions
- You can store functions in data structures
- You can create functions on the fly
- You can create functions that create functions
- Code flexibility
Results In Some Very Useful Features
- Lambda Functions
- Decorators
Example Lambda Function
f = lambda x: x**2
print(f(2))
More Useful Example
num_list = [1,2,3,4,5]
# in just one line
squared = list(map(lambda x: x**2, num_list))
Alt Solution
num_list = [1,2,3,4,5]
# Three lines of code
def f(x):
return x**2
squared = list(map(f, num_list))
Classes
- A blueprint for creating objects
- Objects have properties and methods
- Will not cover in this session
- Requires we cover OOP
- Instead, we will focus on using classes
- Many libraries provide classes
Using Classes
- You must create an instance of the class
- You can then use the methods and properties from the instance
- You can create multiple instances of the same class
- Each instance will have its own properties
- Methods will work on the properties of the instance
- Another way to scope variables
Example of Using Classes
import pandas as pd
df = pd.DataFrame({"col1": [1,2,3]}) # instance
df2 = pd.DataFrame({"A": [4,5,6]}, {"B": [7,8,9]}) # another instance
# calling a method
df.head()
df2.head()
# accessing a property
df.columns
df2.columns
Python Libraries
- Built-in
- Comes with Python
- Third-party
- Installed using pip
Using Libraries
- Import the library
- Read documentation to understand how to use the library
- You can peak into libraries using
dir()andhelp()
- You can peak into libraries using
- Use the library
Example of Using Libraries
import math #simple import
# calling a library function
math.sqrt(4)
# Discovering the library
dir(math)
help(math.sqrt)
Always prefix the function with the library name
Another Example
from math import sqrt # import a specific function
# calling the function
sqrt(4)
Bad Example (Avoid This)
from math import * # import everything
# calling the function
sqrt(4)
Risk of overwriting functions
Example of Aliasing Libraries
import pandas as pd # aliasing
# calling a function/class
pd.DataFrame()
Conventional for most data science libraries
Installing Libraries
$ pip install pandas
Remember:
- This is a shell/terminal command
- If you get an error:
- Try using
pip3instead ofpip - You might need to use
sudoon linux and mac or run terminal as administrator on windows
- Try using
Important Statistical Libraries and Frameworks
- pandas
- numpy
- scipy
- sklearn
- statsmodels
Causal Inference and Bayesian Analysis Libraries
- pymc
- dowhy
- econml
- causalnex
Visualization Libraries
- matplotlib
- seaborn
- plotly
- altair
Network Analysis Libraries
- networkx
- networkit
- graph-tool
- python-igraph
- pydot
Deep Learning Libraries
- tensorflow
- pytorch
- keras
- jax
Natural Language Processing Libraries
- nltk
- spacy
- gensim
- tomotopy
Useful Web/PDF Scraping Libraries
- scrapy
- beautifulsoup
- GROBID
Break
- Resume in 15 minutes
- Make sure pandas library is installed
Introduction to Pandas
Notes
- Recommended to use Jupyter Notebook
- VSCode has a notebook mode
- You can output content of a cell by just typing the variable name
- You can also use the
print()function - Without print you can only see the last line of the cell
- With print you can see multiple lines
- You can also use the
- Copilot and ChatGPT can be very useful
What is Pandas?
- Open source data analysis and manipulation tool
- Built on top of the Python programming language
- Offers data structures and operations for manipulating numerical tables and time series
- Integrates well with other libraries in Python ecosystem
- Name derived from Panel Data
How to install Pandas
$ pip install pandas
Anaconda python comes with Pandas and other libraries pre-installed.
Importing Pandas in your script
import pandas as pd # conventional alias
Setup and configuration typically done at top of script or notebook
Typical Research Setup
# Assumed for all examples
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import statsmodels.api as sm
import sklearn
Pandas Data Structures
Introduction to Series and DataFrame Creating a Series and DataFrame Accessing and modifying data in Series and DataFrame
Series
- One-dimensional labeled array
- Can hold any data type
- Can be created from a list, dictionary, or scalar value
- A combination of a list and a dictionary
- has both numeric and named indices
Creating a Series
# from a list
s = pd.Series([1, 2, 3, 4, 5])
print(s)
# from a dictionary
d = {'a': 1, 'b': 2, 'c': 3}
s = pd.Series(d)
print(s)
# from a scalar value
s = pd.Series(5, index=[0, 1, 2, 3, 4])
print(s)
Accessing and Modifying Data in Series
s = pd.Series([1, 2, 3, 4, 5], index=['a', 'b', 'c', 'd', 'e'])
# Accessing data
print(s['a'])
print(s[0])
# Modifying data
s['a'] = 6
print(s)
DataFrame
- Two-dimensional labeled data structure
- Like a table or spreadsheet
- Can hold any data type
- Can be created from a dictionary, list of dictionaries, or a list of lists
- Each column is a Series of equal length
Creating a DataFrame
# from a dictionary
d = {'col1': [1, 2, 3], 'col2': [4, 5, 6]}
df = pd.DataFrame(d)
print(df)
# from a list of dictionaries
data = [{'a': 1, 'b': 2}, {'a': 3, 'b': 4, 'c': 5}]
df = pd.DataFrame(data)
print(df)
# from a list of lists
data = [[1, 2, 3], [4, 5, 6]]
df = pd.DataFrame(data, columns=['a', 'b', 'c'])
print(df)
Accessing Data in DataFrame
- Can feel confusing at first
- You have multiple options
- Select columns:
[] and . - Select rows:
loc, iloc, and [] - Select rows and columns:
loc, iloc, and [] - Select a cell:
at and iat - There is also query for more complex selection of rows
DataFrames Have an Index
- Used to identify rows
- Can be numeric or named
- Can be set to a column where values become the index
- You can also access rows by number
- Indecis and columns can be hierarchical
- We will not cover this
Selecting Columns
df = pd.DataFrame({'a': [1, 2, 3], 'b': [4, 5, 6], 'c': [7, 8, 9]})
# single column
print(df['a'])
# or
print(df.a)
# multiple columns
print(df[['b', 'a']])
Selecting Rows
df = pd.DataFrame({'a': [1, 2, 3], 'b': [4, 5, 6], 'c': [7, 8, 9]}, index=['x', 'y', 'z'])
# single row by number
print(df.loc[0])
# or by index
print(df.iloc['x'])
# multiple rows (slices)
print(df.iloc[0:1])
# or by index
print(df.loc['x':'y']) # maintains order
Selecting Rows and Columns
df = pd.DataFrame({'a': [1, 2, 3], 'b': [4, 5, 6], 'c': [7, 8, 9]}, index=['x', 'y', 'z'])
# single cell
print(df.at['x', 'a'])
# or
print(df.iat[0, 0])
# multiple cells ()
print(df.loc['x':'y', 'a':'b'])
# or
print(df.iloc[0:2, 0:2])
Notice
- loc and iloc, first argument is for rows and second is for columns
- You can select columns with loc and iloc:
print(df.loc[:, 'a':'b'])
print(df.iloc[:, 0:2])
Conditional Selection of Rows
df = pd.DataFrame({'a': [1, 2, 3], 'b': [4, 5, 6], 'c': [7, 8, 9]}, index=['x', 'y', 'z'])
# single condition
print(df[df['a'] > 1])
# multiple conditions
print(df[(df['a'] > 1) & (df['b'] < 6)])
# or using query
print(df.query('a > 1 and b < 6'))
Loading Data
- You can read data from a file or URL
- Pandas supports many file formats
- CSV, Excel, SQL, JSON, HTML, Stata, SAS, Parquet, Feather, and more
- Use
read_*functions - You might need to install additional libraries for some formats
Loading Data Example
df = pd.read_csv('file.csv')
df = pd.read_excel('file.xlsx')
df = pd.read_sql('SELECT * FROM table', connection)
df = pd.read_json('file.json')
df = pd.read_html('http://example.com/tables.html')
df = pd.read_stata('file.dta')
df = pd.read_sas('file.sas7bdat')
df = pd.read_parquet('file.parquet') # requires pyarrow
Exercise
- Visit this page
- Download any dataset and extract data file
- Make sure data file is in the same directory as your notebook
- Load the dataset into a DataFrame
- Convention is to prefix it with
dffor easy reference - For example,
df_data = pd.read_csv('file.csv')
- Convention is to prefix it with
- Display the dataframe
Data Discovery
- Displaying the first few rows
- Displaying the last few rows
- Displaying the shape
- Displaying the columns
- Displaying the data types
- Displaying the summary statistics
- Displaying the unique values
- Displaying the value counts
Data Set to Use in Analysis
- Load Gapminder dataset from URL
url = "https://raw.githubusercontent.com/resbaz/r-novice-gapminder-files/master/data/gapminder-FiveYearData.csv"
df = pd.read_csv(url)
Data Discovery Example
# Displaying the first few rows
print(df.head())
# Displaying the last few rows
print(df.tail())
# Displaying the shape
print(df.shape)
# Displaying the columns
print(df.columns)
Data Discovery Example
# Displaying the data types
print(df.dtypes)
# Displaying the summary statistics
print(df.describe())
# Displaying the unique values
print(df['country'].unique())
# Displaying the value counts
print(df['country'].value_counts())
Saving Data
- You can save data to a file or database
- Pandas supports many file formats
- CSV, Excel, SQL, JSON, HTML, Stata, SAS, Parquet, Feather, and more
- Use
to_*functions - You might need to install additional libraries for some formats
Saving Data Example
df.to_csv('file.csv')
df.to_excel('file.xlsx')
df.to_sql('table', connection)
df.to_json('file.json')
df.to_html('file.html')
df.to_stata('file.dta')
df.to_sas('file.sas7bdat')
df.to_parquet('file.parquet') # requires pyarrow
Things to Note About Saving and Loading Data
- You can specify the file path
- For arabic text, you might have to specify the encoding
- Likely to be
utf-8
- Likely to be
- You can store or drop the index
Things to Note About Saving and Loading Data
- You can specify data types for each column
- Either during or after loading
- Use dtype argument in
read_*functions - Use
astypemethod in DataFrame
- You can specify date and float formats
Data Cleaning
- Handling missing data
- Removing duplicates
- Renaming and replacing
Missing Data Example
df = pd.DataFrame({
'a': [1, 2, np.nan],
'b': [4, np.nan, 6],
'c': [np.nan, 8, 9]
})
print(df)
# drop rows with missing data
print(df.dropna())
# drop columns with missing data
print(df.dropna(axis=1))
# fill missing data
print(df.fillna(0))
Removing Duplicates Example
df = pd.DataFrame({
'a': [1, 2, 2, 3],
'b': [4, 5, 5, 6],
'c': [7, 8, 8, 9],
})
print(df)
# drop duplicates
print(df.drop_duplicates())
# drop duplicates based on a column
print(df.drop_duplicates(subset=['a']))
Removing Duplicates Example
# display duplicates
print(df[df.duplicated()])
# display duplicates based on a column
print(df[df.duplicated(subset=['a'])])
# Try to print df after each operation
# What do you notice?
DataFrame Immutability
- Operations on a DataFrame do not change the original DataFrame
- You must assign the result to a new variable
- You can use the
inplaceargument to change the original DataFrame- Not recommended
- Can be confusing
- Can be error prone
- Can be slow
- Try dropping duplicates with and without
inplace
Solution
df = pd.DataFrame({
'a': [1, 2, 2, 3],
'b': [4, 5, 5, 6],
'c': [7, 8, 8, 9],
})
print(df)
# drop duplicates, use this with any
# operation that changes the dataframe
df = df.drop_duplicates()
print(df)
# drop duplicates using inplace
df.drop_duplicates(inplace=True)
print(df)
Renaming and Replacing Example
df = pd.DataFrame({
'a': [1, 2, 3], 'b': [4, 5, 6], 'c': [7, 8, 9],})
print(df)
# rename columns
df_renamed = df.rename(columns={'a': 'A', 'b': 'B'})
print(df_renamed)
# alternatively
df.columns = ['A', 'B', 'C']
print(df)
# replace values
df_replaced = df_renamed.replace(1, 100)
print(df_replaced)
Data Analysis
- Descriptive statistics
- Grouping and aggregating data
- Correlation and covariance
Load Gapminder Data
import pandas as pd
url = "https://raw.githubusercontent.com/resbaz/r-novice-gapminder-files/master/data/gapminder-FiveYearData.csv"
df = pd.read_csv(url)
Descriptive Statistics Example
print(df.describe())
# or
print(df['gdpPercap'].mean())
print(df['gdpPercap'].median())
print(df['gdpPercap'].std())
print(df['gdpPercap'].min())
print(df['gdpPercap'].max())
print(df['gdpPercap'].quantile(0.25))
print(df['gdpPercap'].quantile(0.75))
Grouping and Aggregating Data Example
# use gapminder df, it is in df
print(df.groupby('country')['gdpPercap'].mean())
# or
print(df.groupby('country')['gdpPercap'].agg(
['mean', 'median', 'std', 'min', 'max', 'count']))
# or
print(df.groupby('country').agg(
{
'gdpPercap': ['mean', 'median', 'std', 'min', 'max', 'count'],
'lifeExp': ['mean', 'median', 'std', 'min', 'max', 'count'],
}
))
Aggregating Without Reshaping DataFrames
- You can use the
transformmethod to aggregate without reshaping the DataFrame
df['gdpPercap_mean'] = df.groupby('country')['gdpPercap'].transform('mean')
print(df)
- Notice how we assign the result to a new column
Correlation and Covariance Example
- The following will give an error, can you fix it?
print(df.corr())
print(df.cov())
Solution
# You need to select the numeric columns
corr_matrix = df[['year', 'pop', 'lifeExp', 'gdpPercap']].corr()
cov_matrix = df[['year', 'pop', 'lifeExp', 'gdpPercap']].cov()
# To correlate specific columns
df['lifeExp'].corr(df['gdpPercap'])
df['lifeExp'].cov(df['gdpPercap'])
Data Visualization
- Pandas has some built-in plotting capabilities
- You can also use matplotlib and seaborn for more advanced plots
- You can also use plotly for interactive plots
- You can also use altair for declarative plots (similar to ggplot2 in R)
- We will cover only matplotlib and seaborn
Pandas Plotting Example
# just choose the column and call the plot method
df['gdpPercap'].plot()
# You can choose the kind of plot
df['gdpPercap'].plot(kind='hist')
# You can also use the plot method of the DataFrame
df.plot(x='year', y='gdpPercap', kind='scatter')
Plot Kinds Supported by Pandas
- line
- bar
- barh
- hist
- box
- kde
Plot Kinds Supported by Pandas
- density
- area
- pie
- scatter
- hexbin
- Each has its own restrictions and requirements
Commulative Sum Example
df['gdpPercap'].cumsum().plot()
Matplotlib
- The most popular plotting library for Python
- Provides a MATLAB-like interface
- Can be used to create complex, publication-quality plots
Steps To Create a Plot
- Create a figure
- Create one or more subplots
- Plot data on the subplots
- Customize the plot
- Save the plot
- Show the plot
Example
import matplotlib.pyplot as plt
# Create a figure
fig = plt.figure()
# Create a subplot
ax = fig.add_subplot(111)
# this creates a 1x1 grid of subplots and returns the first one
# alternatively, you can use fig.add_subplot(1, 1, 1)
# Plot data on the subplot
ax.plot([1, 2, 3, 4], [10, 20, 25, 30])
# x-axis is the first list and y-axis is the second list
# replace lists with pandas series or numpy arrays
# Customize the plot (optional)
ax.set_xlabel('x-axis')
ax.set_ylabel('y-axis')
ax.set_title('Title')
# Save the plot (optional)
plt.savefig('plot.png')
# Show the plot
plt.show()
4 Subplot Matplotlib Example
fig, axs = plt.subplots(2, 2)
axs[0, 0].plot(df['year'], df['gdpPercap'])
axs[0, 0].set_title('GDP Per Capita')
axs[0, 1].plot(df['year'], df['lifeExp'])
axs[0, 1].set_title('Life Expectancy')
axs[1, 0].plot(df['year'], df['pop'])
axs[1, 0].set_title('Population')
axs[1, 1].plot(df['year'], df['gdpPercap'] * df['pop'])
axs[1, 1].set_title('GDP')
plt.show()
Problem
Plots were a mess, can you fix them?
Solution
- Select a subset of the data for a specific country
df_kenya = df[df['country'] == 'Kenya']
# or use query
df_kenya = df.query('country == "Kenya"')
# replace df with df_kenya in the previous example
Seaborn
- Built on top of matplotlib
- Provides a high-level interface for drawing attractive and informative statistical graphics
- Provides a variety of plot types, color palettes, and themes
- You might find it easier and defaults more appealing than matplotlib
- Importing seaborn will also set the matplotlib and pandas defaults
- Grouping is easier
Seaborn Example
import seaborn as sns
# Code is similar to matplotlib
# but you can also use the sns function
sns.lineplot(x='year', y='gdpPercap', data=df)
Seaborn Plot Types
- Has many types to list here
- Discover them from documentation at:
Seaborn Example
# scatter plot grouped by continent
sns.scatterplot(x='year', y='gdpPercap', data=df, hue='continent')
# box plot grouped by continent
sns.boxplot(x='continent', y='gdpPercap', data=df)
# violin plot grouped by continent
sns.violinplot(x='continent', y='gdpPercap', data=df)
# pair plot
sns.pairplot(df[['gdpPercap', 'lifeExp', 'pop']])
# heatmap
sns.heatmap(df[['year', 'pop', 'lifeExp', 'gdpPercap']].corr())
# count plot
sns.countplot(x='continent', data=df)
Seaborn Themes
- Seaborn is highly customizable
- You can set the theme, color palette, and context
- Redraw the plot after setting the theme
sns.set_theme(style='whitegrid') # alternatives include 'darkgrid', 'white',
# 'dark', 'ticks'
sns.set_palette('pastel') # alternatives include 'deep', 'muted',
# 'bright', 'dark', 'colorblind'
sns.set_context('talk') # this sets the font size for talk
# you can also use 'paper', 'notebook', 'poster'
Reshaping Data
- Get familiar with the concepts of tidy data
- Make data tidy
- You meld to convert wide to long format
- Useful for analysis
- You pivot to convert long to wide format
- Useful for summaries
- You meld to convert wide to long format
- Concatenate and merge data
Tidy Data
- Requirements:
- Each variable forms a column
- Each observation forms a row
- Each type of observational unit forms a table
Tidy Data
- 3rd normal form in databases
- Can be long or wide
- Many tools and libraries assume data is tidy
- Especially for visualization
Data Example
| Name | treatmenta | treatmentb |
|---|---|---|
| John Smith | — | 2 |
| Jane Doe | 16 | 11 |
| Mary Johnson | 3 | 1 |
-Is this tidy?
source: (Wickham 2014)
Another Data Example
| Name | John Smith | Jane Doe | Mary Johnson |
|---|---|---|---|
| treatmenta | — | 16 | 3 |
| treatmentb | 2 | 11 | 1 |
-Is this tidy?
source: (Wickham 2014)
Yet Another Data Example
| Name | Treatment | Result |
|---|---|---|
| John Smith | a | — |
| Jane Doe | a | 16 |
| Mary Johnson | a | 3 |
| John Smith | b | 2 |
| Jane Doe | b | 11 |
| Mary Johnson | b | 1 |
-Is this tidy?
source: (Wickham 2014)
Converting from Wide to Long Format
df = pd.DataFrame({
'Name': ['John Smith', 'Jane Doe', 'Mary Johnson'],
'treatmenta': [None, 16, 3],
'treatmentb': [2, 11, 1]})
# Melt the DataFrame to long format
df_long = df.melt(
id_vars=['Name'],
var_name='Treatment',
value_name='Result')
print(df_long)
Melted Data
| Name | Treatment | Result |
|---|---|---|
| John Smith | treatmenta | — |
| Jane Doe | treatmenta | 16 |
| Mary Johnson | treatmenta | 3 |
| John Smith | treatmentb | 2 |
| Jane Doe | treatmentb | 11 |
| Mary Johnson | treatmentb | 1 |
Lets fix treatment to be a and b
Solution
# just remove the word treatment from the data
tidy_df['Treatment'] = tidy_df['Treatment'].str.replace('treatment', '')
What Happened Here?
df.melt(id_vars=['Name'], var_name='Treatment', value_name='Result')
id_varsspecifies the columns to keep as is (identifier variables)var_namespecifies the name of the new column that contains the wide column namesvalue_namespecifies the name of the new column that contains the values from the wide columns
Converting from Long to Wide Format
df_long.pivot(index='Name', columns='Treatment', values='Result')
Pivot vs Pivot Table
pivotis a reshaping method- Does not aggregate data
- Requires unique index/column pairs
- Otherwise, you will get a
ValueError
- Otherwise, you will get a
pivot_tableis a reshaping method- Aggregates data
- Does not require unique index/column pairs
- Has more arguments
- Used to create a summary table
Pivot Table Example
df = pd.DataFrame({'A': ['foo', 'foo', 'foo', 'bar', 'bar', 'bar'],
'B': ['one', 'one', 'two', 'two', 'one', 'one'],
'C': ['small', 'large', 'large', 'small', 'small', 'large'],
'D': [1, 2, 2, 3, 3, 4]})
print(df)
# pivot table
print(df.pivot_table(index='A', columns='B', values='D'))
# More arguments
print(df.pivot_table(index='A',
columns=['B', 'C'], values='D', aggfunc='sum',
fill_value=0, margins=True, margins_name='Total'))
Statistical Analysis with Statsmodels
Introduction to Statsmodels
- Statistical modeling and testing
- Integrates well with pandas
- Provides R-style model specification
- Comprehensive output that you can interact with
- Documentation can be found here
Available Models
- Various linear models
- Time series analysis
- Much more
- Examples is another great resource for learning
OLS Example
import statsmodels.api as sm
data = sm.datasets.longley.load_pandas()
print(data.data) # whole dataset
print(data.endog) # endogenous variable
print(data.exog) # exogenous variables
print(data.endog_name)
print(data.exog_name)
res = sm.OLS(data.endog, data.exog).fit()
print(res.summary())
Goodness Of Fit Example
import statsmodels.api as sm
import matplotlib.pyplot as plt
fig = sm.graphics.qqplot(res.resid, line='q')
plt.show()
GLM Example (R-style formula)
import statsmodels.api as sm
import statsmodels.formula.api as smf
data = sm.datasets.get_rdataset('epil', package='MASS').data
print(data)
mod = smf.glm(
"y ~ age + trt + base",
data,
family=sm.families.Poisson()
)
res = mod.fit()
print(res.summary())
# check for overdispersion
if res.pearson_chi2 / res.df_resid > 1:
print('We have overdispersion')
else:
print('No evidence to suggest overdispersion')
Exploring Statsmodels
- Documentation is extensive
- Examples are very useful
- Check them out here
Machine Learning with Scikit-learn
What is Scikit-learn?
- Simple and efficient tools for ML and statistical modeling
- Can be used for classification, regression, clustering, dimensionality reduction, and more
- Built on NumPy, SciPy, and matplotlib
- Open source, commercially usable - BSD license
- Documentation can be found here
- Check also common pitfalls and best practices here
Scikit-learn FlowChart
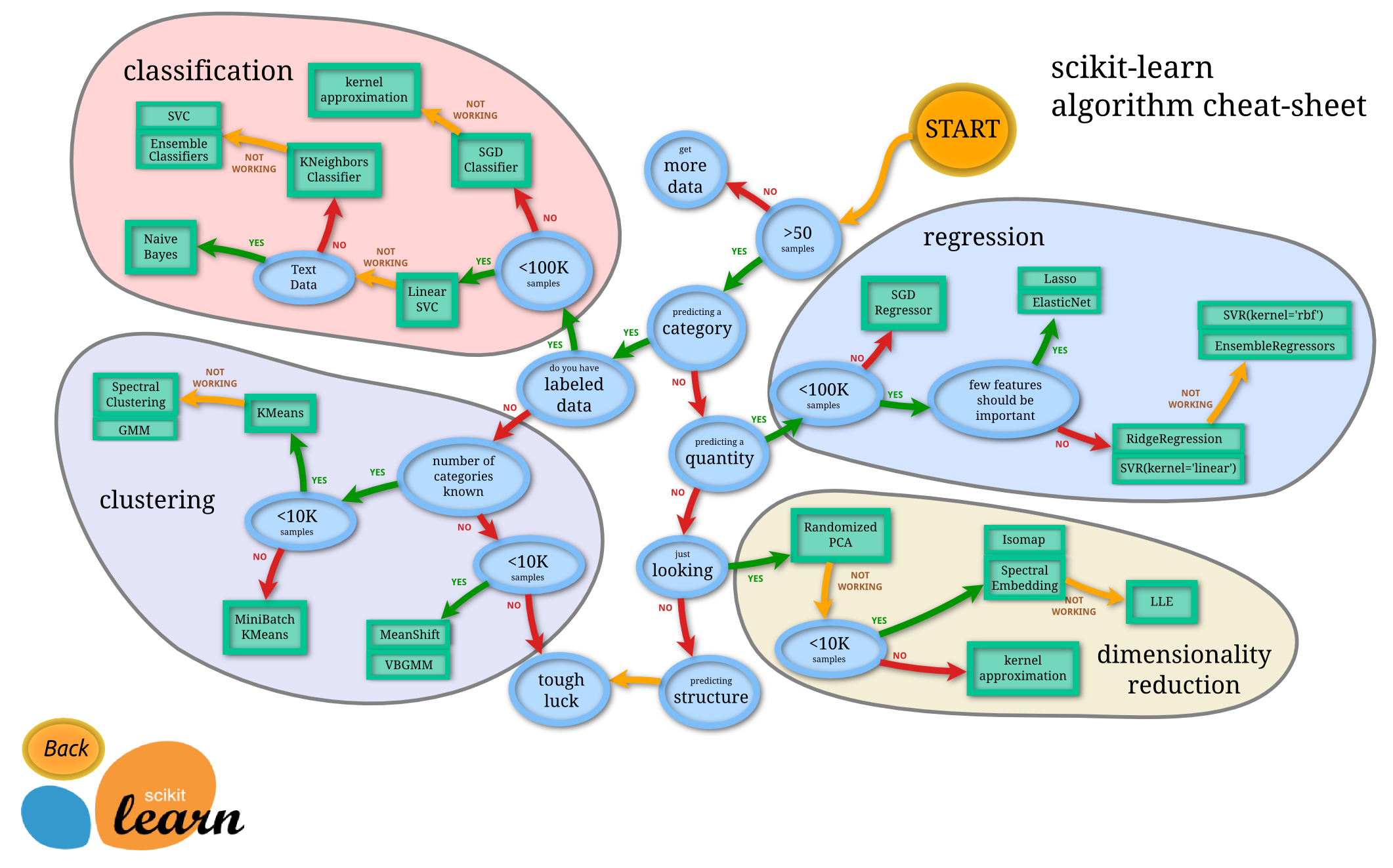
So Much To Learn
- Little time to cover everything
- Someone has probably solved your problem in Python
- You can find a library, documentation, tutorials, and a discord community for almost anything
- Have fun exploring!
Reproducible Research Documents
Using Quarto
What is Quarto?
- Open Source technical publishing system
- Designed for reproducible research
- Language agnostic
- Supports Python, R, Julia, and more
- IDE agnostic
- Supports Jupyter, RStudio, VSCode, and more
- Embeds code, data, results, references, and narrative
What is Quarto?
- Utilizes Markdown
- Can be used to produce documents, reports, presentations, books, and websites
- Supports multiple output formats
- HTML, PDF, Word, Jupyter Notebooks, LaTex and more
- Facilitates collaboration and sharing
- Best when used with version control systems (e.g, Git)
- Documentation can be found here
What is Markdown?
Let’s Start Using Quarto
- Install Quarto
- Create a new Quarto project
- In VSCode press Shift+Ctrl+p for command panel
- Type
Quartoand selectCreate New Project
- Create a manuscript project
- Choose directory to store project and give it a name
Let’s Start Using Quarto
- Explore configuration, qmd file options, and authoring
- Explore Quarto guide and try Getting started guide
Explore Following Features
- Previewing documents in different formats
- Explore markdown
- Embedding code and options
- Embed variables inline
- Draw plots and diagrams
- Use references and citations
- Use mermaid diagrams
Final Thoughts
- Use Python to become proficient
- https://software-carpentry.org/lessons/
Thank You
Course Material
https://malmarz.netlify.app/en/courses/pyintro/
Dont’ Forget